For upload to M-CHiPS mergeblocks will convert your .gpr files into a separate file for each channel that can be uploaded into your database. From the plethora of .gpr file's columns, it extracts the mean of pixels for each spot along with the mean of pixels for each spot's background. This is common practice (also in other labs). It is safe also when label intensity is inhomogeniously distributied over the spot area (e.g. "donuts"). Instead of invoking the function mergeblocks, you can use mergeblocks_meanxpixels, mergeblocks_median, or mergeblocks_medianxpixels, which are described below. They work exactly as mergeblocks does, except that instead of the mean they extract what follows the underscore in their names. For further explanation, ask Kurt. For to judge, what applies to your chips, ask an experienced wet-lab chipper. If in doubt, use the default (i.e. mergeblocks).
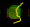
If your spots are not completely homogenous, stick to the mean. A common example of an inhomogenous pixel distribution within one spot is the "donut" shape (see Fig.). Here, a considerable amount of the label is concentrated in the spot margins. The majority of the spots (in the middle part) shows a much lower intensity, thus misleading the median, whereas the mean correctly represents the total amount of bound label:
Only if your spots look homogenous - it may be a good idea to ask someone who had a look at many slides before - , you may successfully apply the median to protect from dust contaminations. Here, in all cases where a dust particle affects less than half of the pixels of a spot, the majority of its pixels will yield a median completely unaffected by the ultra-high fluorescence of the dust particle.
Likewise, if you use a single-channel platform, multiplying with individual pixel numbers makes no difference when spot sizes are constant.
Multiplying with the number of pixels may represent the sum of bound label better than just averaging intensities (may median or mean). But at the risk of overdoing things:
Make shure that adaption to small spots is limited by a minimum spot size !!! When confronted with background - level spots but not bound by a minimum spot size, Genepix tends to home in on one or two pixels, whose intensity level will be more or less due to chance alone (instead of reflecting the background level). While this is bad enough for two-channel data, a pixel number of one or two in this case leads to a factor up to 80-fold lower than for the cases in which for some reason Genepix decides to recognize a normally-sized background-level spot. Thus, huge artifact ratios will definitely mess up your analysis, when forgetting to specify a minimum spot size!
When you intend to multiply with the number of pixels, do not correct any spot by making it smaller in Genepix! Because the number of pixels will be smaller. While this makes a lot of sense to avoid e.g. dust or other unwanted spot areas when the spotsize is not taken into account, this is not an option here.