M-CHiPS News
Multi-Conditional Hybridization Intensity Processing System
Latest developments:
- Live-DVD:
- Knoppix-based M-CHiPS live DVD for demonstration purposes
- Runs on every PC (without installing anything) or newer Mac
- Just boot from DVD and start testing
- Including demonstration data: single-channel, two-channel, and 2D-gel data sets
- Access of own data possible (only from within DKFZ or if stored on local hard drive, but thus you could indeed use M-CHiPS on your next train or flight)
- Full analysis capabilities (comprising all functions available in Oct 2007 except those for upload and annotation)
 )"-button pressed while starting the
computer, klick on Windows - Icon, press Enter.
)"-button pressed while starting the
computer, klick on Windows - Icon, press Enter. Knoppix deals with almost any hardware component. If the system freezes after "PCMCIA found, starting cardmgr...", restart by entering "knoppix nopcmcia" on the bottom of the blue screen after "boot:".
- M-CHiPS update in 2007:
- More intuitive GUI
- Enhanced connectivity for plugging own functions (e.g. Matlab, R, Perl) and external tools (SAM Macro, GenMapp)
- Normalization: lo(w)ess and VSN added
- Filtering: permutation - based (SAM) and non-permutation-based (eBayes) p-values added
- Other functions added
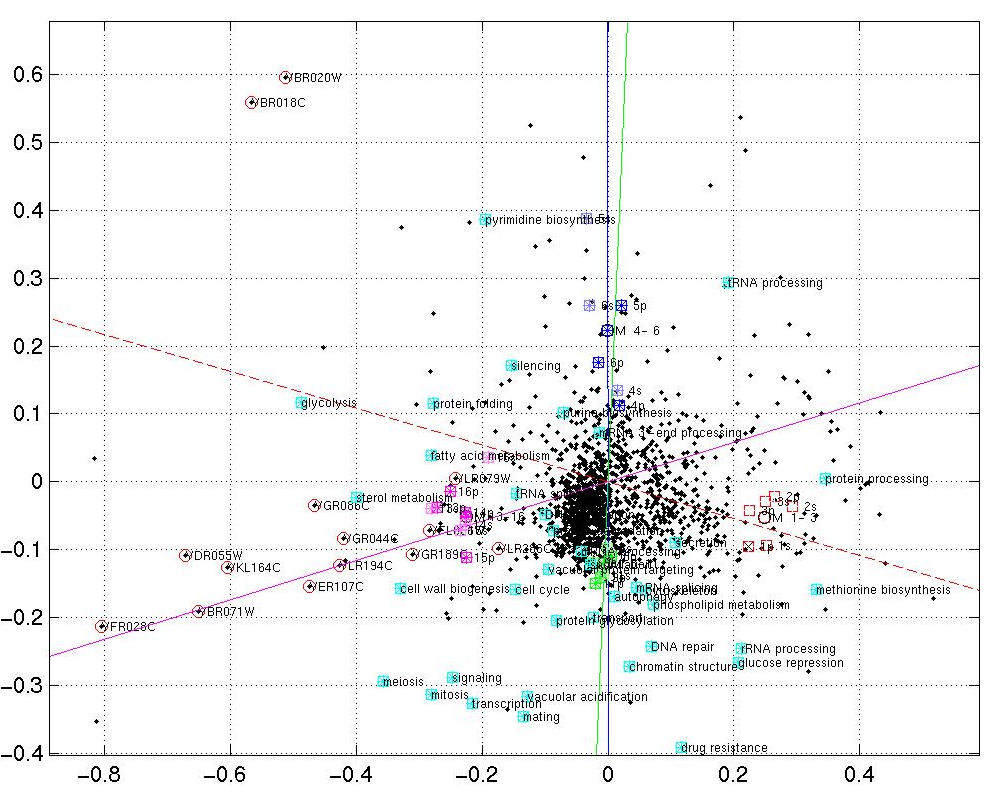
- Gene function goes correspondence analysis :-)
To identify gene functions involved in transcription patterns recorded by microarray hybridization, we simultaneously visualize genes, measurements (i.e. hybridizations) and gene function categories. Content and color code of the following screenshots are identical to [1] Fig. 2 exept for the additional gene function categories (cyan boxes or spheres). They are aimed to provide a small glance at method development in progress and should be viewed as preliminary results.
- Integrated high-end 3D viewer
Many thanks to Dieter Finkenzeller for interfacing his excellent 3D viewer to the package. cl-Vishn uses OpenGL for seemless rotation/zoom and communicates with M-CHIPS via TCP/IP sockets. Among other features it provides independent color-selection for arbitrary sets of genes and hybridizations as well as text labels and easy selection of genesets.
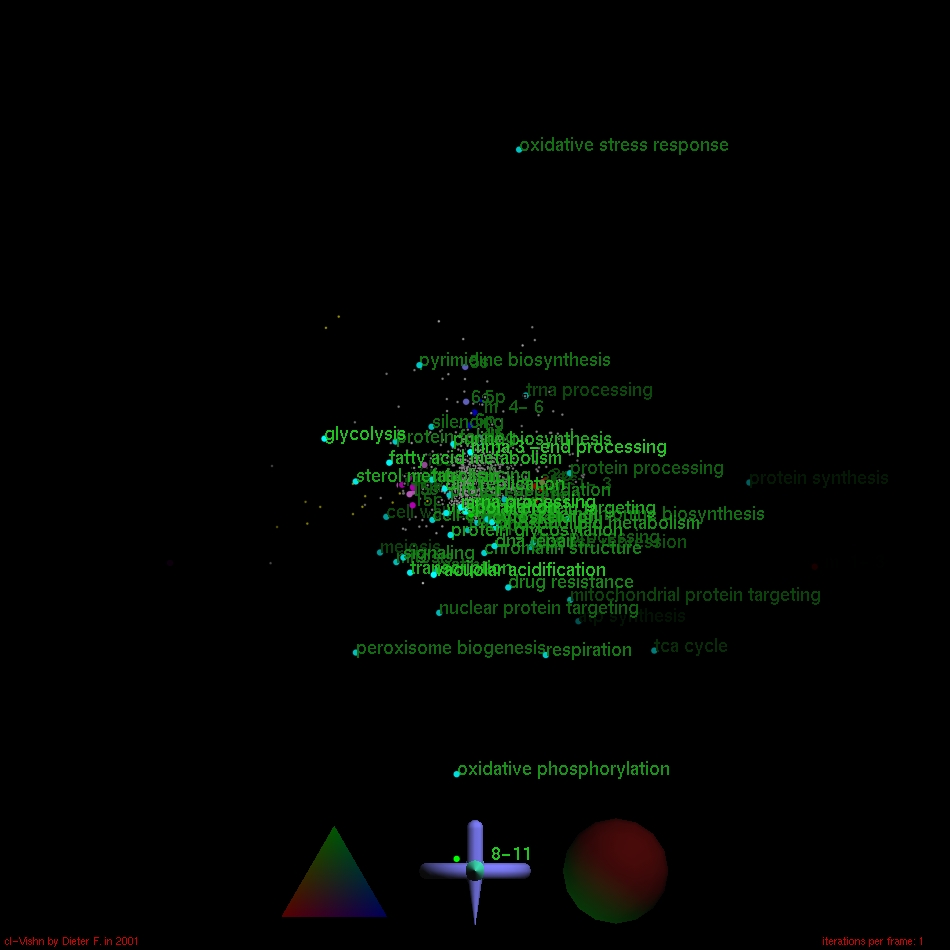
Best viewed with stereo glasses 8-)