| Manual |
| Getting Started |
| Starting M-chips |
| Loading data |
| Normalizing the data |
| Filtering the data |
| Exploring the data |
The mchips server has access to the IPAG storage.
TMT6x
1. Please provide a _CID.mgf (with isotope fitting) and a _HCD.mgf (without isotope fitting: activate "Single Peak Window" in distiller).2. Put both files to put_MGFs_here.
3. Grant read access to ipag(Unix Group\ipag) for both files: rightklick on the file, properties, security, ipag(Unix Group\ipag), grant read access, ok. In case there is no security tab you need to disable simple file sharing: click Start, My Computer, Tools Menu, Folder Options, View tab, Advanced Settings, clear the "use simple file sharing (recommended)" check box.
4. Open a Nx connection to mchips, but instead of typing mchips, type cid_hcd2.pl<enter>.This produces a lot of files. It also shows you the distribution of TMT ion masses for each channel (blue) as well as their intensities (red). The last plot shows the distribution of distances between masses. Here, you need to zoom in (e.g. rectangle select area by keeping left mouse button pressed, zoom out with right mouse button) in order to see that actually two gaussian curves exist for all distances but 5 Da. Press Ok and close to close all windows and produce more files in put_MGFs_here
5. Use the _cut_cid.mgf file for an MS/MS Ions Search.
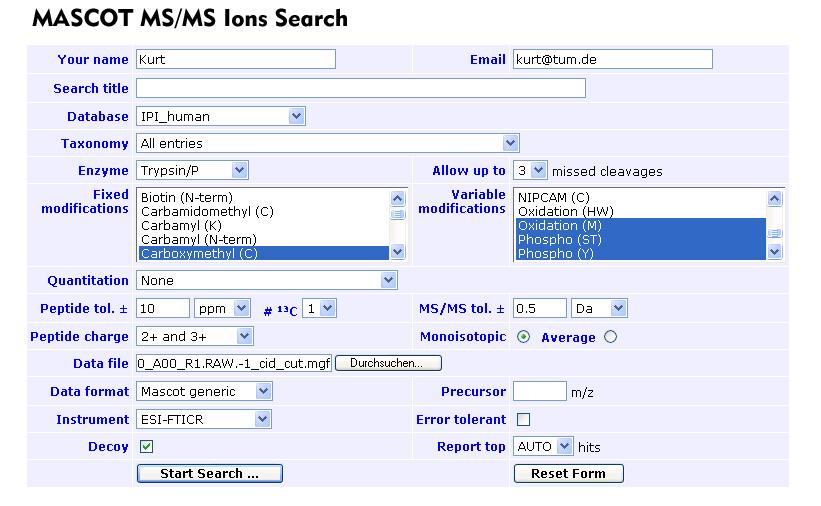
Search parameters can be stored on your machine as a cookie. After the search, click export (Error message: In the adressbar delete the "i" in "idata"), Export format: pepXML, p<0.05, select Description and Mass, deselect pI. Save the .xml file in put_MGFs_here (takes a while, too) and grant read access to ipag(Unix Group\ipag).
If it is too big to export via webserver, do this.
6. On mchips (yellow text window) type pid.pl <enter> which will parse the .xml file.
7. On mchips type the following two lines, each followed by <enter>:
cd put_MGFs_here
mchips
8. Click Data Import, Upload Experiment, (evt. the table family to upload to),yellow textwindow: type n<enter>, click define new experiment and provide an experiment name in the text window. In case all six channels are to be uploaded in their usual sequence, the number of conditions excl. control is 5, not monochannel, n for the next question (because there are more non-control measurements than control measurements), 6x 1 (there is only one measurement for each condition), yes (Input filenames from file), yes (correct sequence, CONTROL FIRST!), 6x 1 (to affiliate the same run 1 to all measurements), n after run 1 was sucessfully entered everywhere, y to start the upload.
Upload may take a while but it does not need any further action from the user side. After importing the quantification data (files .quant131 through .quant126) and the identification data (.id file) it will display
-- SUCCESSFUL COMPLETION :-) --
Data are uploaded in all-or-none fashion, i.e. when there was a problem any time during the upload, the database remains completely unchanged.
2010-06-25