Intro
|
PhyResSE is not a black box!
Genotype and Resistance are based on well-characterized SNPs collected in
one
list ("Resi-List-<version>.txt"). You can
As it will change over time, it is important to record its version at the time of analysis. Export (see below) records the current version of the SNP list in the header (along with other critical metadata such as the current version of the program as well as the one of the reference genome). However, the header will comprise only the SNP list version, but not its content. Please download the SNP list along with your results to document exactly which SNPs your analysis has been based on. | 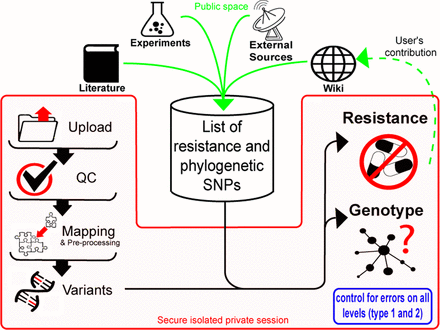 |
Handling
The first table lists variants already reported to be resistance associated. The + icon of the quality column reveals whether or not considerably strict quality criteria are met. The first (position) column displays the nucleotide distribution at each position by mouse-over.It also links to a simple viewer detailing all contributing reads, color-coded according to base (top panel) and mapping (bottom) quality. Links by other columns provide further detail about genes and publications involved. More columns can be added by means of the "Skipped columns" + icon (likewise columns can be skipped by clicking the according x icons).
For safety, variants at any position reported to be resistance associated are listed, even when the called nucleotide differs from the ones already recorded. Differing nucleotides are depicted in grey. The plus icon at the table bottom reveals all nucleotides recorded in our list.
Variants not recorded are listed in a second table (rightmost column)
when
a) located in the CDS of a resistance-related gene if not representing a
silent point mutation.
b) Variants up to 300bp upstream of the gene start are also listed if not
located within the CDS of another gene (for the time being, that includes
hypothetical
proteins).
The second table furthermore reports any position within the CDS of a resistance related gene, the poor (or excess) coverage of which may lead to a type 2 error (failure to discover an existing resistance).
Export Table data can be taken home as described. While both tables have an export button located nearby, each button exports the content of both tables at the same time into one spreatsheet.

Computational Steps
(Warning: Wee little nerdlings only. Log-likelyhood increases with the age of a tree? Best stop reading here.)- no other programs invoked -